


|
|
Home |
DynOmics 1.0 |
Tutorials |
Theory |
References |
iGNM 2.0 |
ANM 2.0 |
Computational Biology |
PITT site
|
Theory
Hitting/Commute Time – Intramolecular Communication
The Markovian stochastic model of information diffusion has been developed for exploring the inter-residue communication in proteins (1). The first passage time in Markovian process, which is the average time (number of steps) for residue/node i to transfer the “message” from node i to j for the first time, is defined as hitting time H(j,i) and H(i,i) = 0. The process is controlled by transition probabilities for the passage of information across the nodes (residues). Specifically, the atomic contact affinity mij between pair of nodes i-j defines the conditional probability which consist of the transition matrix M = {mij}.
![]() (1)
(1)
where
![]() (2)
(2)
and
![]() (3)
(3)
A = {aij} and D = diag{dij} are the affinity and degree matrices respectively. Nii is the number of heavy atom contacts between residues i and j based on a cutoff distance of rc = 4 Å. Ni and Nj are the number of heavy atoms in residue i and j respectively. Notably, the Kirchhoff matrix (combinatorial Laplacian) G in GNM theory can be defined as
G = D - A (4)
Based on this, the hitting time (information-theoretic quantities) was bridged to the GNM-defined intrinsic structural dynamics (statistical mechanical theory) of proteins.
Residues vi and vj are termed as the broadcaster (perturbation site) and receiver (response site) respectively. The passage from vi to vj can be performed in two stages: (i) from vi to its directly connected neighbor residues (termed as intermediate residue vk) that is one step away (∑nk=1 mki = 1); (ii) succeeded by probabilistic passages from vk to the final destination vj. An efficient recursive formula can be derived as
![]() (5)
(5)
Then we are able to calculate the hitting time between two any nodes from the above equation using a self-consistent method. The commute time between vi and vj is termed as
C(j,i)= H(j,i) + H(i,j) = C(i,j) (6)
The commute time matrix C = {C(j,i)} is symmetric while the hitting time matric H = H(j,i) is not.
Technically, the “fundamental matrix” technique can also be used to calculate these quantities (2).
Based on the bridging between G and A in Eq. 4, we obtained
![]() (7)
(7)
where G-1 is the pseudo-inversion of G. The elements in G-1 are to the intrinsic dynamics of residues in GNM theory. We can rewrite the Eq. 7 as
![]() (8)
(8)
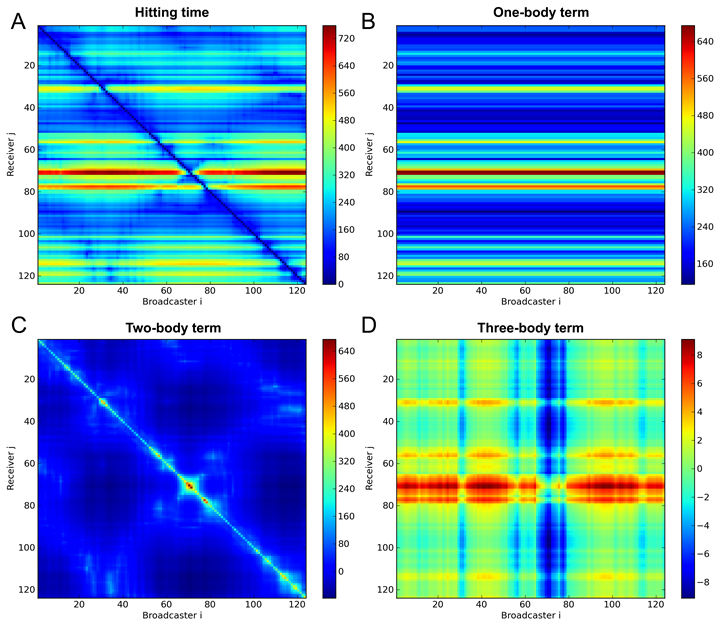
Figure 1. Hitting time (A) of Phospholipase A2 (PDB id: 1BK9) and the decomposed hitting time: one-body term (B), two-body (C) term and three-body term (D). The figures were reproduced from (1).
The detailed derivation of Eq. 7 can be seen in (1). Based on the understanding of this equation, we can tell that the hitting time can be decomposed into three terms: one-body term, which is the mean-square fluctuation of the response site (<∆rjT∆rj>); two-body term, which depends on the cross-correlations between residues vi and vj (-<∆rjT∆ri>); and the three-body term, which depends on the cross-correlations between intermediate residue vk and residues vi and vj (<∆rkT∆ri> - <∆rkT∆rj>). Take the protein Phospholipase A2 (PDB id: 1BK9) as an example, the hitting time (Fig.1 A) of protein Phospholipase A2 was decomposed into three terms (Fig.1 B-D). The one-body term (Fig.1 B) plays a dominant role the two-body term (Fig.1 C) has the visible effect in specific regions. While the effect of the three body term (Fig.1 D) is negligibly small.
In the example of Phospholipase A2, the average Receiver (response site) hitting times of the three catalytic residues H49, Y52 and D99 are in a smallest level (sensitive; efficient to receive signals).
Reference
1. Chennubhotla, C. and Bahar, I. (2007) Signal Propagation in Proteins and Relation to Equilibrium Fluctuations. PLoS Comp. Biol., 3, e172.
2. Norris, J.R. (1997) Markov chains. Cambridge (United Kingdom): Cambridge University Press.